Malaria Detection
Problem Statement
Malaria is a disease caused by parasites transmitted through bites from female mosquitoes. Symptoms of malaria include fever, vomiting, and even death in severe cases. There were an estimated 228 million malaria cases worldwide in 2018, with an estimated death toll of 405,000 people. Malaria has a strong presence in developing countries, especially within regions of Africa where 93% and 94% of total malaria cases and deaths occurred, respectively. The World Health Organization (WHO) suggests that “prompt diagnosis and treatment is the most effective way to prevent a mild case of malaria from developing into severe disease and death." However, the WHO reports that a considerable proportion of children who exhibited symptoms of a fever (me dian 36%) did not seek any medical attention whatsoever, listing poor access to healthcare and ignorance of malaria symptoms as notable contributing factors.
The diagnosis of malaria from blood smears usually requires a highly skilled technician to count and classify red blood cells. The task of counting infected blood cells and calculating infection rates are better left to computers than to humans, especially since most developing countries already have microscopic technologies available but lack the required resources for training and educating technicians to interpret blood smears properly. To alleviate the impact of healthcare worker shortages in developing countries, we created a machine learning-based software to help automate these two tasks. With our approach, operating microscopes and performing blood smear stains are still required of clinicians, but the technical burden of interpreting images and counting infected cells are offloaded to computers. In turn, places that once lacked trained technicians can now benefit from more screening sites due to the reduced need for specialization. Clinicians can focus on treating patients rather than deciphering blood smears and allow technology to streamline the malaria diagnosis process.
Implementation
Training and Results
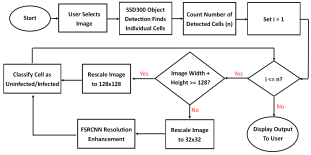
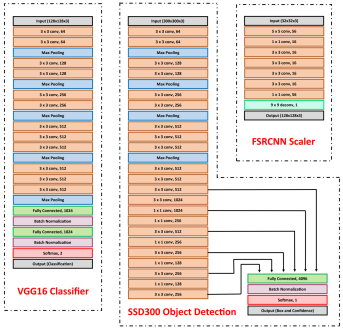
Our model consists of three neural networks that process the blood smear sequentially: an object detection model, a resolution enhancement model, and a cell classification model.
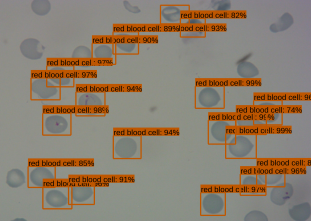
The object detection model consists of a pre-trained single shot multibox detector with an image input size of 300x300 (SSD300). The SSD300 object detection model is able to detect the presence of red blood cells with an average precision of 90.4% when the Intersection over Union (IoU) is 0.50 for all area sizes
The resolution enhancement model is a faster super-resolution convolutional neural network (FSRCNN) with four mapping layers, a high-resolution dimension of 56, and a low-resolution dimension of 16. The FSRCNN model takes in low resolution 32x32 images and artificially enhances it to high resolution 128x128 images to maximize the amount of detail to be analyzed by subsequent classification. The best performing FSRCNN has a peak signal-to-noise ratio (PSNR) of 30.79 and a mean squared errror (MSE) of 54.66. In contrast, the traditional method of bicubic interpolation yielded a PSNR of 24.10 and a MSE of 254.67. In addition, the FSRCNN-derived images are classified more accurately than the raw low resolution images or bicubic interpolated images in the finalized CNN classification model

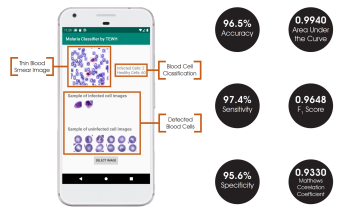
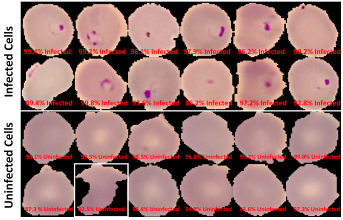
The final classification model is a variant of the VGG16 neural network, with batch normalization layers implement after each dense layer prior to passing through the activation function. Each dense layer consisted of 1024 nodes instead of the standard 4096 nodes to minimize computational burden. Our final VGG16 classifier model had an accuracy of 96.5%, sensitivity of 97.4%, specificity of 95.6%, and AUC of 0.9940. The full architectural details of all three models is shown below.
App Development
While all models were developed and trained with the TensorFlow and Keras packages, the final model deployments are subsequently converted into a .tflite file that allows the models to be run on TensorFlow Lite. TensorFlow Lite is an open-source platform focused on on-device model inference. Unlike previously reported studies that utilize phone apps for model prediction, this allows the models to run directly on the Android-based smartphones rather than relying on cloud-based computing resources.
The Android app takes in a user-selected image of a Giemsa-stained thin blood smear. Then, the SSD300 model isolates individual images of the red blood cells and discards images of white blood cells. if the individual images are of low resolution, the FSRCNN model enhances the resolution. Finally, each red blood cell image is classified by the VGG16 model to count the number of uninfected and infected red blood cells, as shown in Figure 2. Lastly, the file size of the SSD300, FSRCNN, and VGG16 models are 95 MB, 5 MB, and 20 MB, respectively, which is lower than the 200 MB file size limit. A summary of our data pipeline is depicted below.